|
|
| Model | FSSP generic growth model |
| References |
Dalgaard, P. (2009). Modelling of
microbial growth. Bulletin of the International Dairy Federation, 433 45-57. Mejlholm, O. and Dalgaard. P. (2009). Development and validation of an extensive growth and growth boundary model for Listeria monocytogenes in lightly preserved and ready-to-eat shrimp. J. Food Prot. 70, 2132-2143.
Mejlholm, O, Dalgaard, P. (2013).
Development and
validation of an extensive growth and growth boundary model for
psychrotolerant Lactobacillus
spp. in seafood and
meat products.
Int. J.
Food Microbiol.
167, 244-260.
|
| Primary growth model | Logistic model with
delay |
| Secondary growth model | Simplified cardinal parameter type model |
| Environmental parameters in model | Temperature, atmosphere (CO2), water phase salt/aw, pH, smoke components/phenol, nitrite and organic acids in water phase of product (acetic acids, benzoic acid, citric acid, diacetate, lactic acid and sorbic acid) |
|
This generic model is a highly flexible tool to predict growth of various microorganisms in different foods. The FSSP generic growth model allows users to define values of 12 environmental parameters (See the FSSP dialog box and eqn 1 below). In addition users can specify a reference maximum specific growth rate (Áref, 1/h) for each model and a relative lag time (RLT value. In ths way the FSSP generic growth model can predict responses of most microorganism/food combinnations and this flexible tool has many applications. The FSSP generic growth model can for example be used to:
|
|
How to use the FSSP generic model tool: To start using the FSSP generic model tool you must first 'Add model' or 'Import model' and then Select growth model (sse FSSP dialog box below).
|
|
Fig. 1. FSSP dialog box for the Generic growth model. |
|
When pressing the 'Add model' bottom the dialog box below appears and allow users to (i) modify the indicated default paramter values and (ii) name and save the new model. Saved models can be Exported to an XML-file. This file can be send to other users of FSSP and they can easily Import, Select and used this model. When a model is selected in FSSP it can be modified by pressing the 'Edit model' bottom and then changes can be saved under the same or under a new model name. FSSP Generic growth models are highly flexible but if a model is used for a fresh food product without salt, without smoke, without nitrite and/or without added organic acids then these model terms can easily be inactivated by applying the relevant boxes in the 'Not in use' column (See dialog box belowe). |
|
|
|
Fig. 2. 'Add model' dialog box used to enter new parameter values and to save a new model. |
|
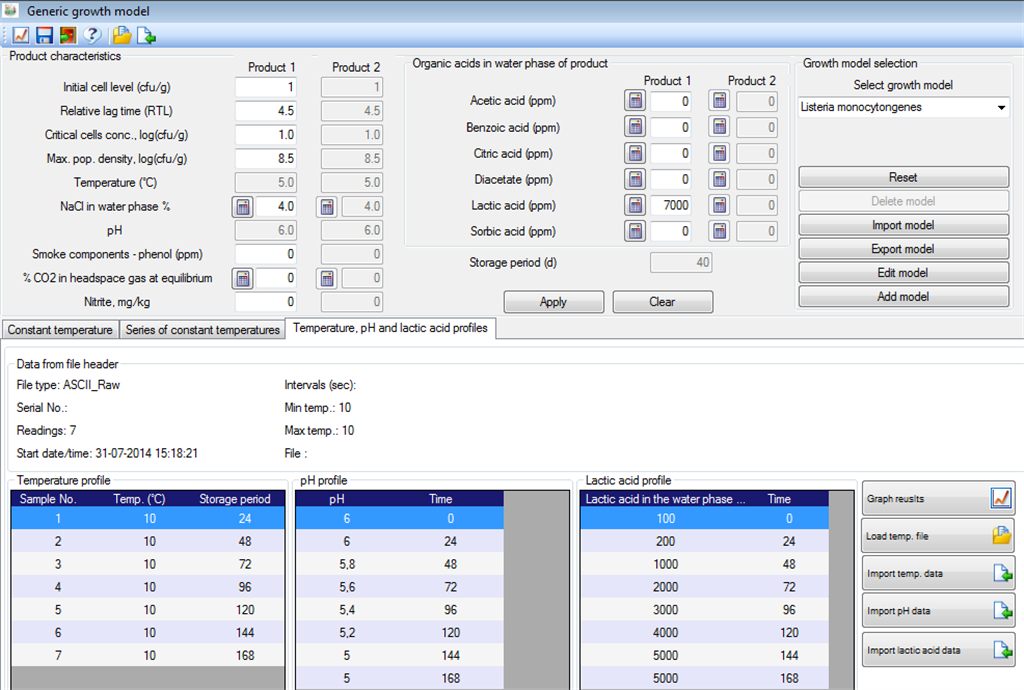
Like for other models in FSSP the Generic growth model can provide predictions at constant and dynamic temperature store conditions. Figure 1 shows an example where growth is predicted at a constant temperature of 8.0░C. Figure 1 also showns how Generic growth models can be used with relative lag time (RLT) values selected by the user. In addition to dynamic storage temperaures FSSP can predict the effect of changing pH and changing lactic acid concentrations on growth resonses. Figure 3 and Figure 4 below show how pH and lactic acid profile data are imported by using the FSSP raw data importer. Fugure 5 below shows how the predicted growth (red curve) is dampend when pH (purple curve) decreases below ca. 5.5 and the concentration of lactic acid (green curve) increases to more then ca. 2000 ppm. |
 |
|
Fig. 3. Import of temperature, pH and lactic acid profiles by usinf the FSSP raw data
importer. |
 |
|
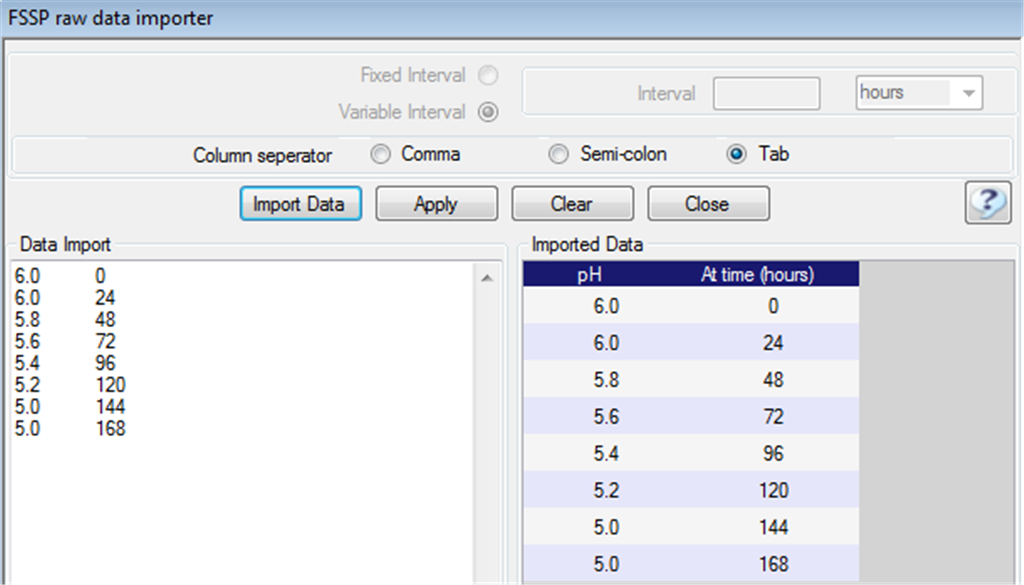
Fig. 4. The FSSP raw data importer allow data to be transfered
too FSSP from spreadsheets by copy and paste. |
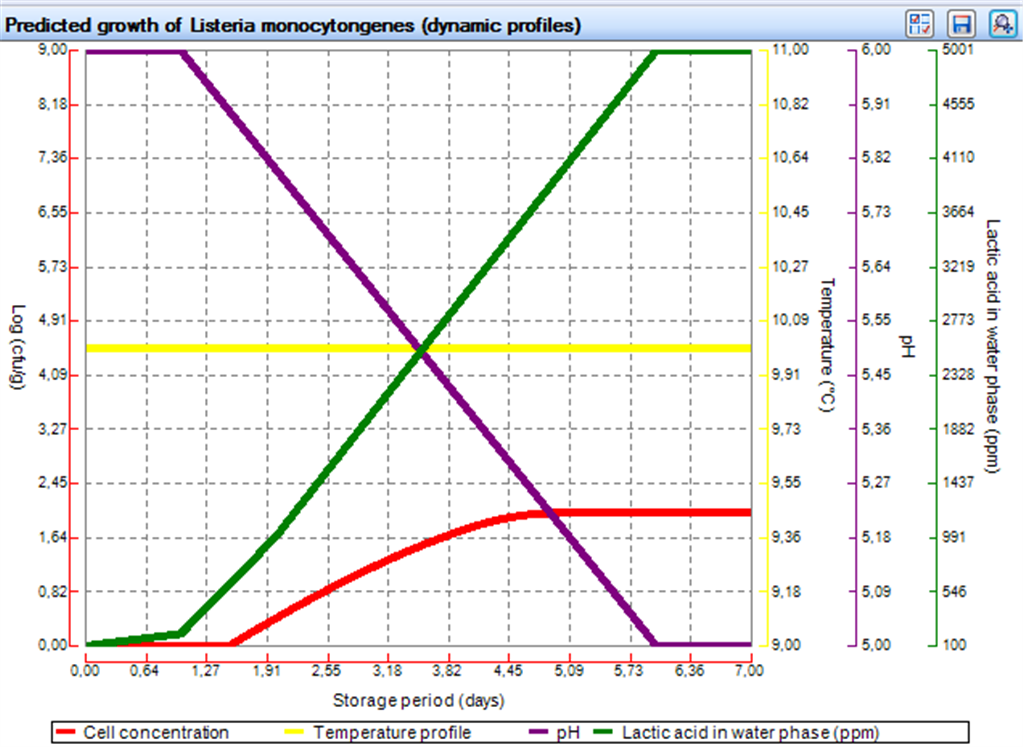 |
| Fig. 4. FSSP Generic growth model output window showing how growth is dammpend when pH decreases and the lactiac concentration oncreases. |
|
|
|
| Eqn. 1. Logistic model with delay. |
|
|
|
| Eqn. 2. Secondary growth and growth boundary model used by the FSSP Generic growth model tool. |
|
|
|
|
| Fig. 5. FSSP dialog box showing how MIC values for undissociated organic acids and model terms for each organis acid is indicatd with appropriate values of n1 and n2.. |
|
|
|
|